One of my favorite things to do is craft graphical user interface (GUI) applications.
It can be really fulfilling when you write an app that satifies some purpose or
need. There's a lot you can do and the possibilities seem almost endless.
A friend of mine who majored in micro-biology, in undergrad, had to write a .fasta
file reader. Fasta format is just a standardized way in which DNA sequences are
stored in a file. She only had to write a command line interface program to read
and display the file. I asked her if she would send me the project. She obliged
my request and I did make the command line fasta program. I realized it was a
really good opportunity to write it into a GUI application. It was one of the first
GUI apps I ever wrote—I learned a lot while writing it.
There are three prominent modules in Python for writing GUI apps; Kivy, PyQt and
Tkinter. I started out with Kivy but have come to realize that PyQt is far
superior—at least for writing desktop applications. I havn't used Tkinter much at
all.
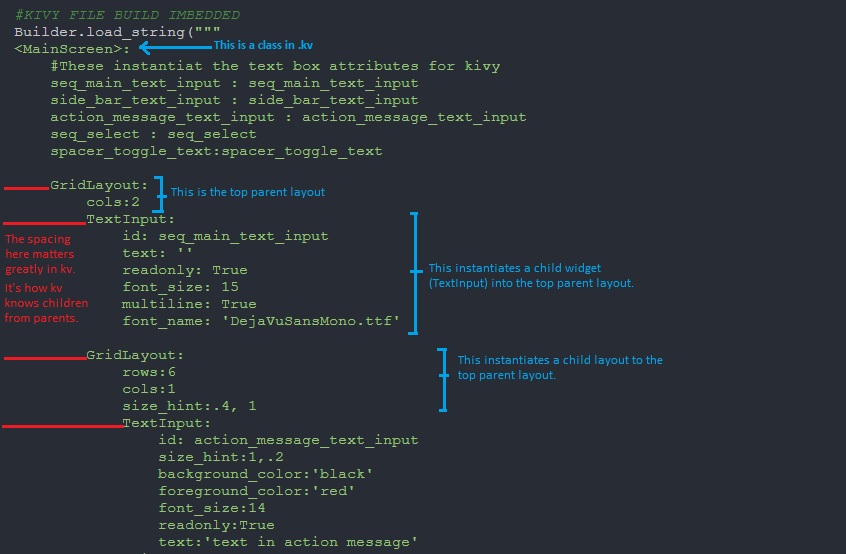
The first version of the DNA Sequence Viewer that I wrote used Kivy. I then re-wrote
the code using PyQt5. For me this was good exersize becuase both modules do have
different strengths and weaknesses. Kivy has a really unique look—it's somewhat
intended for mobile applications. Whilst PyQt is a very solid desktop application
module.
PyQt Version
PyQt5 Version Demo Video:
PyQt5 Imports
#IMPORTS
from PyQt5.QtWidgets import *
from PyQt5.QtGui import *
from PyQt5.QtCore import *
import sys
from collections import Counter
import pandas as pd
import matplotlib.pyplot as plt
Brief review of PyQt5 methods
STEPS 1 and 2:
One of the first tasks after importing the necessary modules and instantiating
a class for your window is to instantiate a layout—somewhat like a master
layout that all other layouts will reside in.
#IMPORTS
from PyQt5.QtWidgets import *
from PyQt5.QtGui import *
from PyQt5.QtCore import *
import sys
from collections import Counter
import pandas as pd
import matplotlib.pyplot as plt
class WindowClassOne(QWidget):
def __init__(self):
super(WindowClassOne,self).__init__()
#SET THE DEFAULT FONT FOR APP
self.setFont(QFont('consolas',11))
#SET DEFUALT MINIMUM SIZE OF WINDOW
self.setMinimumSize(900,900)
#INSTANTIATE AN OUTER LAYOUT (master layout); STEP 1
self.outerLayout = QHBoxLayout()
#ADD THE MAIN WINDOWS OUTER LAYOUT (MASTER LAYOUT); STEP 2
self.setLayout(self.outerLayout)
if __name__ == "__main__":
app = QApplication(sys.argv)
app.setStyle('Fusion')
window = WindowClassOne()
window.show()
sys.exit(app.exec_())
When you run the above it outputs a blank window. This is where you add layouts and widgets.
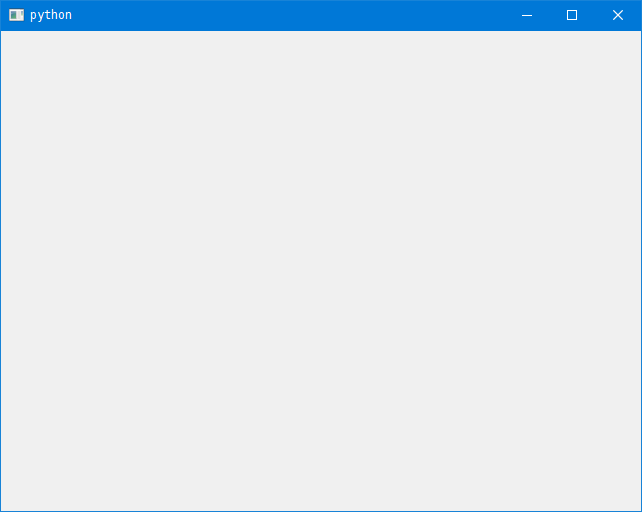
Steps 3 and 4:
Add a sub-layouts and/or widgets to the main layout (here named self.outerLayout).
This next step depends on what your app requires. For my purposes here
I need to add a left and right sublayout so I'll instantiat them and call
them to the main layout.
#IMPORTS
from PyQt5.QtWidgets import *
from PyQt5.QtGui import *
from PyQt5.QtCore import *
import sys
from collections import Counter
import pandas as pd
import matplotlib.pyplot as plt
class WindowClassOne(QWidget):
def __init__(self):
super(WindowClassOne,self).__init__()
#SET THE DEFAULT FONT FOR APP
self.setFont(QFont('consolas',11))
#SET DEFUALT MINIMUM SIZE OF WINDOW
self.setMinimumSize(900,900)
#INSTANTIATE AN OUTER LAYOUT (master layout); STEP 1
self.outerLayout = QHBoxLayout()
#ADD THE MAIN WINDOWS OUTER LAYOUT (MASTER LAYOUT); STEP 2
self.setLayout(self.outerLayout)
#INSTANTIATE LEFT AND RIGHT LAYOUTS (sub layouts); STEP 3
self.leftLayout = QGridLayout()
self.rightLayout = QGridLayout()
self.subformlayout_1 = QFormLayout()
self.subrightlayout = QGridLayout()
#CALL THE INNER LAYOUTS INTO THE OUTER LAYOUT (MASTER LAYOUT); STEP 4
self.outerLayout.addLayout(self.leftLayout, 7) #the integer indicates respective stretch
self.outerLayout.addLayout(self.rightLayout, 2) #the integer indicates respective stretch
if __name__ == "__main__":
app = QApplication(sys.argv)
app.setStyle('Fusion')
window = WindowClassOne()
window.show()
sys.exit(app.exec_())
The above will still yield a blank window but the layouts are created and
called.
Steps 5 and 6: Instantiate and call widgets to the layouts.
#IMPORTS
from PyQt5.QtWidgets import *
from PyQt5.QtGui import *
from PyQt5.QtCore import *
import sys
from collections import Counter
import pandas as pd
import matplotlib.pyplot as plt
class WindowClassOne(QWidget):
def __init__(self):
super(WindowClassOne,self).__init__()
#SET THE DEFAULT FONT FOR APP
self.setFont(QFont('consolas',11))
#SET DEFUALT MINIMUM SIZE OF WINDOW
self.setMinimumSize(900,900)
#INSTANTIATE AN OUTER LAYOUT (master layout); STEP 1
self.outerLayout = QHBoxLayout()
#ADD THE MAIN WINDOWS OUTER LAYOUT (MASTER LAYOUT); STEP 2
self.setLayout(self.outerLayout)
#INSTANTIATE LEFT AND RIGHT LAYOUTS (sub layouts); STEP 3
self.leftLayout = QGridLayout()
self.rightLayout = QGridLayout()
self.subformlayout_1 = QFormLayout()
self.subrightlayout = QGridLayout()
#CALL THE INNER LAYOUTS INTO THE OUTER LAYOUT (MASTER LAYOUT); STEP 4
self.outerLayout.addLayout(self.leftLayout, 7) #the integer indicates respective stretch
self.outerLayout.addLayout(self.rightLayout, 2) #the integer indicates respective stretch
#INSTANTIATE WIDGETS (added in order) STEP 5
self.left_layout_textedit = QTextEdit('LEFT LAYOUT HERE...', readOnly=True)
self.right_layout_textedit = QTextEdit('RIGHT LAYOUT HERE...', readOnly=True)
self.some_button_example = QPushButton('SOME BUTTON')
#CALL THE WIDGETS TO THE LEFT AND RIGHT LAYOUTS; STEP 6
#you have to declare where the widget goes in the gridlayout here
#so syntax; SomeLayout.addWidget(SomeWidget, gridCoord_y, gridCoord_x)
#span multi syntax; SomeLayout.addWidget(SomeWidget, gridCoord_y, gridCoord_x, gridCoord_y2, gridCoord_x2)
self.leftLayout.addWidget(self.left_layout_textedit, 0, 0, 1, 0)
self.rightLayout.addWidget(self.right_layout_textedit, 1, 0, 1, 0)
self.rightLayout.addWidget(self.some_button_example, 0, 0, 1, 0)
if __name__ == "__main__":
app = QApplication(sys.argv)
app.setStyle('Fusion')
window = WindowClassOne()
window.show()
sys.exit(app.exec_())
When you run the code after adding steps 5 and 6 we can see the widgets in the app. But the button wont do anything until we; write a function and bind it to the button.
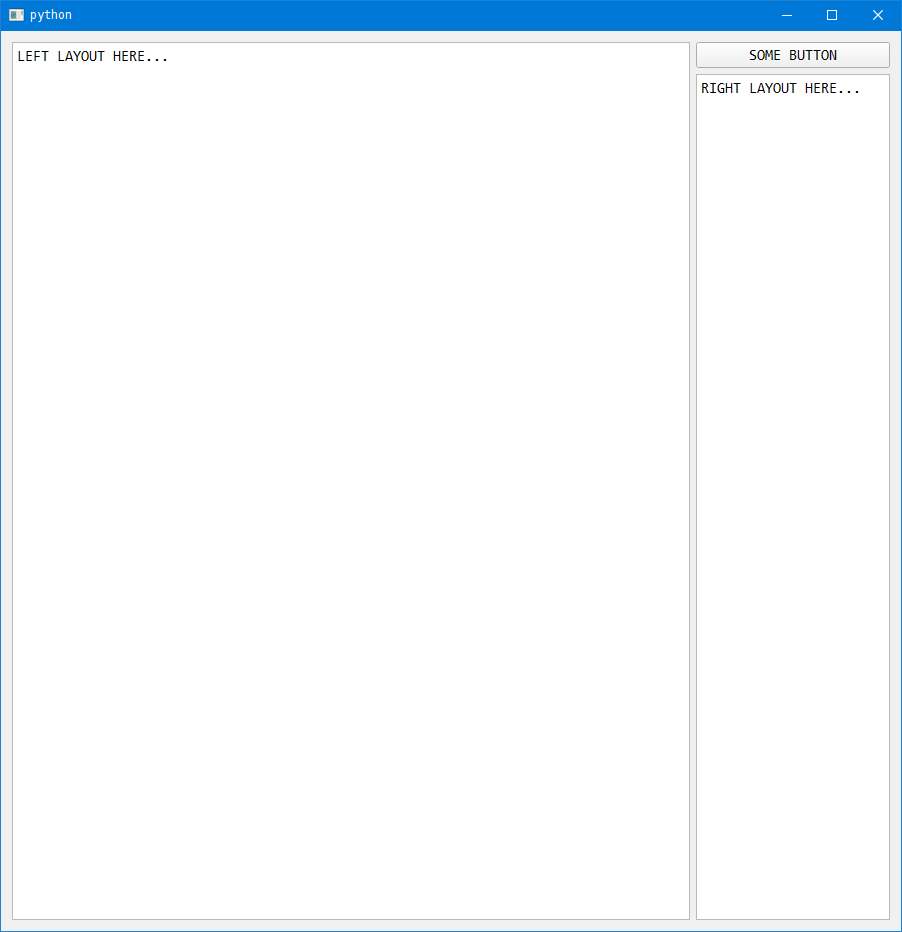
Steps 7 and 8: Write a function and bind (connect) it to the button.
#IMPORTS
from PyQt5.QtWidgets import *
from PyQt5.QtGui import *
from PyQt5.QtCore import *
import sys
from collections import Counter
import pandas as pd
import matplotlib.pyplot as plt
class WindowClassOne(QWidget):
def __init__(self):
super(WindowClassOne,self).__init__()
#set the default font for app
self.setFont(QFont('consolas',11))
#SET DEFUALT MINIMUM SIZE OF WINDOW
self.setMinimumSize(900,900)
#INSTANTIATE AN OUTER LAYOUT (master layout); STEP 1
self.outerLayout = QHBoxLayout()
#ADD THE MAIN WINDOWS OUTER LAYOUT (MASTER LAYOUT); STEP 2
self.setLayout(self.outerLayout)
#INSTANTIATE LEFT AND RIGHT LAYOUTS (sub layouts); STEP 3
self.leftLayout = QGridLayout()
self.rightLayout = QGridLayout()
self.subformlayout_1 = QFormLayout()
self.subrightlayout = QGridLayout()
#CALL THE INNER LAYOUTS INTO THE OUTER LAYOUT (MASTER LAYOUT); STEP 4
self.outerLayout.addLayout(self.leftLayout, 7) #the integer indicates respective stretch
self.outerLayout.addLayout(self.rightLayout, 2) #the integer indicates respective stretch
#INSTANTIATE WIDGETS (added in order) STEP 5
self.left_layout_textedit = QTextEdit('LEFT LAYOUT HERE...', readOnly=True)
self.right_layout_textedit = QTextEdit('RIGHT LAYOUT HERE...', readOnly=True)
self.some_button_example = QPushButton('SOME BUTTON')
#CALL THE WIDGETS TO THE LEFT AND RIGHT LAYOUTS; STEP 6
#you have to declare where the widget goes in the gridlayout here
#so syntax; SomeLayout.addWidget(SomeWidget, gridCoord_y, gridCoord_x)
#span multi syntax; SomeLayout.addWidget(SomeWidget, gridCoord_y, gridCoord_x, gridCoord_y2, gridCoord_x2)
self.leftLayout.addWidget(self.left_layout_textedit, 0, 0, 1, 0)
self.rightLayout.addWidget(self.right_layout_textedit, 1, 0, 1, 0)
self.rightLayout.addWidget(self.some_button_example, 0, 0, 1, 0)
#WRITE A FUNCTION THAT DOES SOMETHING; STEP 7
def something_to_do(self):
self.left_layout_textedit.append('This text is appended to the LEFT when the button is pushed')
self.right_layout_textedit.append('This text is appended to the RIGHT when the button is pushed')
#BIND (CONNECT) THE BUTTON TO THE FUNCTION; STEP 8
self.some_button_example.clicked.connect(lambda checked: something_to_do(self))
if __name__ == "__main__":
app = QApplication(sys.argv)
app.setStyle('Fusion')
window = WindowClassOne()
window.show()
sys.exit(app.exec_())
Now when we run the program and click the button, whatever is in the function (something_to_do) is executed.
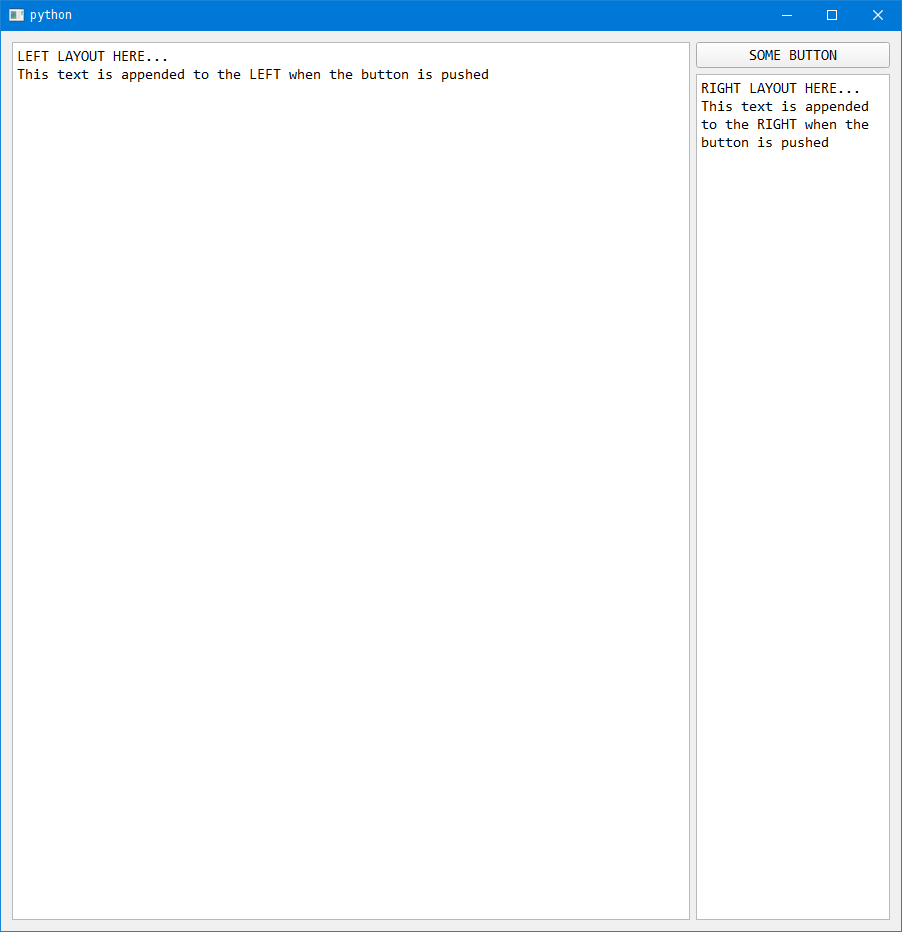
Depending on what program you are trying to write and what you want it to do this process can get rather complex. For now I'm going to leave the brief overview at this.
Full PyQt5 ANG DNA Parser V5_1: The program I wrote to parse and
view fasta files pretty much uses everything above—although with more layouts,
widgets, and functions.
This program isn't perfect and there is a lot I want to do to it. It works
as intended but I do need to fix some of the try/except statements in some
of the functions. I also want to add an data log, exporter, and more graphing
functions. For now I need to move on to other things.
I've done my best to comment heavily, here is the full code:
# -*- coding: utf-8 -*-
"""
@author: ANG
"""
#IMPORTS
from PyQt5.QtWidgets import *
from PyQt5.QtGui import *
from PyQt5.QtCore import *
import sys
from collections import Counter
import pandas as pd
import matplotlib.pyplot as plt
#TO CATCH PYtoEXE ERRORS
def except_hooky(type, value, traceback, oldhook=sys.excepthook):
'''
Had a slight issue converting this file to an exe. This function freezes the
output so I could read the TraceBack error. Without this it closed too fast
for me to read it.
'''
oldhook(type, value, traceback)
input("Press RETURN. ")
sys.excepthook = except_hooky
class WindowClassOne(QWidget):
def __init__(self):
super(WindowClassOne,self).__init__()
#set the default font for app
self.setFont(QFont('consolas',11))
#INSTANTIATE AN OUTER LAYOUT (master layout) #STEP 1
self.outerLayout = QHBoxLayout()
#INSTANTIATE LEFT AND RIGHT LAYOUTS (sub layouts) #STEP 3
self.leftLayout = QGridLayout()
self.rightLayout = QGridLayout()
self.subformlayout_1 = QFormLayout()
self.subrightlayout = QGridLayout()
#INSTANTIATE LEFTLAYOUT WIDGETS #STEP 5
self.seq_main_text_input = QTextEdit('', readOnly=True)
self.seq_main_text_input.moveCursor(1, 0)
#INSTANTIATE LAYOUT WIDGETS #STEP 6
#ACTION MESSAGE
self.action_message_text_input = QTextEdit('Load a fasta file...', readOnly=True)
self.action_message_text_input.setMaximumHeight(60)
#SIDE BAR STATS TEXT
self.side_bar_text_input = QTextEdit('Load a fasta file... ', readOnly=True)
#LOAD FASTA BUTTON
self.load_fasta_button = QPushButton('Load Fasta File')
#SELECT SEQ BUTTON
self.select_seq_button = QPushButton('Select Sequence')
#SELECT SEQ INPUT START
self.select_seq_text_box = QLineEdit('')
self.select_seq_text_box.setMaximumWidth(100)
#SELECT SEQ INPUT STOP
self.select_seq_text_box_stop = QLineEdit('Stop')
#SPACER TOGGLE LABEL
self.spacer_tog_label = QLabel('Spacer Toggle')
#SPACER TOGGLE CHECKBOX
self.spacer_tog_checkbox = QCheckBox()
#SHOW SEQ BY 50 BUTTON
self.show_seq_data_by_50 = QPushButton('Show Seq Data By 50')
#SHOW SEQ BY 100 BUTTON
self.show_seq_data_by_100 = QPushButton('Show Seq Data By 100')
#PLOT GC DATA BUTTON
self.plot_gc_button = QPushButton('Plot GC Data')
#ADD WIDGETS TO LEFT LAYOUT (added in order) STEP 7
self.leftLayout.addWidget(self.seq_main_text_input,0,3,1,1)
#CALL WIDGETS TO RIGHT LAYOUT (added in order) STEP 8
#you have to declare where the widget goes in the gridlayout here
#so syntax; SomeLayout.addWidget(SomeWidget, gridCoord_y, gridCoord_x)
#span multi syntax; SomeLayout.addWidget(SomeWidget, gridCoord_y, gridCoord_x, gridCoord_y2, gridCoord_x2)
self.rightLayout.addWidget(self.action_message_text_input, 0, 0, 1, 0)
self.rightLayout.addWidget(self.side_bar_text_input, 1, 0, 1, 1)
self.rightLayout.addWidget(self.load_fasta_button, 2, 0, 1, 1)
#CALL THE INNER LAYOUTS INTO THE OUTER LAYOUT (MASTER LAYOUT) #STEP 4
self.outerLayout.addLayout(self.leftLayout, 7) #the integer indicates respective stretch
self.outerLayout.addLayout(self.rightLayout, 2) #the integer indicates respective stretch
#CALL THE INNER LAYOUTS (SUB-LAYOUTS)
self.rightLayout.addLayout(self.subformlayout_1, 3, 0)
self.rightLayout.addLayout(self.subrightlayout, 4, 0,)
#INSTANTIATE THE SUB FORM LAYOUTS
self.subformlayout_1.addRow(self.select_seq_button, self.select_seq_text_box, )
self.subformlayout_1.addRow(self.spacer_tog_label, self.spacer_tog_checkbox)
#ADD TO SUBRIGHT LAYOUT
self.subrightlayout.addWidget(self.show_seq_data_by_50, 1, 0, 1, 1)
self.subrightlayout.addWidget(self.show_seq_data_by_100, 1, 1, 1, 1)
self.subrightlayout.addWidget(self.plot_gc_button, 2, 0, 1, 1)
#SET SOME STYLES
#SET STYLE OF WHOLE APP
self.setStyleSheet("background-color : #ADC6C5 ; height:30px ; font: consolas ")
#INSTANTIATE FUNCTION TO DO SOMETHING (CHANGE THE ACTION TEXT BOX TEXT) #STEP 9
def changer(self):
self.action_message_text_input.setText('text has been edited')
self.seq_main_text_input.insertPlainText('\nYou need to load a .fasta...')
print('button pressed')
def load_fasta_alert(self):
self.load_fasta_alert = QMessageBox()
self.load_fasta_alert.setWindowTitle('Alert!!!')
self.load_fasta_alert.setText('You need to load a .fasta file...')
self.load_fasta_alert.exec_()
#FILE LOADER FUNCTION (very simple file loader)
def load_file(self):
'''
Intended to load the file as an object. Essentially this function
takes the text in the .fasta file as a string. Separates it every greater
than operator. Then splits the titles that start with greater than operator
and the actual sequence data into to separate lists with the same
index. In retrospect it may have been more efficient to use a dictionary
but the code functioned as intended so I left it as lists. [Works as intended].
'''
try:
#OPEN_FILE RETURNS A TUPLE
filename = QFileDialog.getOpenFileName(None, 'Open file', './', 'FASTA Files (*.fasta)')
txt = open(filename[0], 'r')
with txt:
self.data = txt.read()
self.filename = filename[0].split('/')
print(self.filename)
#####Segment the data into respective [data_keys] [data_vals] lists#####
genes = self.data.split('>')[1:]
seqdic = {}
for gene in genes:
gene = gene.strip().split('\n')
seqdic['>'+gene[0]] = ''.join(gene[1:])
seqdic_keys_lst = list(seqdic.keys())
seqdic_vals_lst = list(seqdic.values())
self.original_data_lists = seqdic_vals_lst
#REMOVE A STRING VALUE FROM A LIST OF STRINGS
self.data_keys = seqdic_keys_lst
self.data_vals = seqdic_vals_lst
#print(self.data_keys, self.data_vals)#Printed for debugging
self.spacer = 'Off'
#SIDE BAR DISPLAY
self.side_bar_text_input.setText('')
self.side_bar_text_1 = 'There are {0} sequences in {1}\n'.format(len(self.data_keys), self.filename[-1])
self.side_bar_text_2 = 'Sequence selection range: 0 - {}\n'.format(len(self.data_keys)-1)
self.side_bar_text_3 = '\nSequence Selected: '
side_bar_str_lst = [self.side_bar_text_1, self.side_bar_text_2, self.side_bar_text_3]
for i in side_bar_str_lst:
self.side_bar_text_input.insertPlainText(i)
self.action_message_text_input.setText('')
self.action_message_text_input.insertPlainText('Fasta file has loaded...\nChoose a sequence...')
except:
self.action_message_text_input.setPlainText('File Load canceled...')
def reselect_seq_alert(self):
'''
This function calls an alert box that tells the user their input into
the select sequence text input was invalid.
'''
self.load_fasta_alert = QMessageBox()
self.load_fasta_alert.setWindowTitle('Alert!!')
self.load_fasta_alert.setText('The sequence you selected is out of range.\nOr you need to input an integer.\nre-select a sequence')
self.load_fasta_alert.exec_()
def load_a_fasta_alert(self):
'''
This function calls an alert box that tells the user they need to
load a .fasta file if one is not already loaded.
'''
self.load_fasta_alert = QMessageBox()
self.load_fasta_alert.setWindowTitle('Alert!!')
self.load_fasta_alert.setText('User needs to load a .fasta')
self.load_fasta_alert.exec_()
def select_seq(self):
'''
This function assigns the text the user input as the selected seq.
It then checks that the number input is in the range of the index of
data keys. If it is not it alerts the user in the upper right action
message box.
'''
try:
self.seq_selected = self.select_seq_text_box.text()
self.seq_keys_nums = [str(i) for i in range(0, len(self.data_keys))]
if self.seq_selected not in self.seq_keys_nums:
self.action_message_text_input.setText('The sequence you selected is out of range.\nOr you need to input an integer.')
else:
pass
except:
pass
def select_seq_ana(self):
'''
Intended to parse the statistics for the selected sequence and display
them in the right_layout text box.
'''
try:
file_data_2 = self.data.split('>')[1:] #converts file_data to list of lists
seq_selected_idx = int(self.seq_selected)
scoped_data = file_data_2[seq_selected_idx].split('\n')[0:] #creates a list of all data split by \n
self.heading_title = scoped_data[0]
counter_1 = Counter()
for i in self.original_data_lists[int(self.seq_selected)]:
counter_1[i] += 1
counter_1 = dict(counter_1)
self.counter_at = self.original_data_lists[int(self.seq_selected)].count('AT')
self.counter_gc = self.original_data_lists[int(self.seq_selected)].count('GC')
self.counter_cn = self.original_data_lists[int(self.seq_selected)].count('N')
self.n_counts_header = '\nNucleotide counts: \n'
n_counts_lst = []
for k, v in counter_1.items():
n_counts = '{} = {}'.format(k, str(v))
n_counts_lst.append(n_counts)
self.n_counts_lst_str = '\n'.join(n_counts_lst)
self.total = self.counter_at + self.counter_gc
stat_at = self.counter_at/self.total
stat_gc = self.counter_gc/self.total
self.at_stat = 'AT = {} : AT content = {}'.format(str(self.counter_at), str(round(stat_at, 5)))
self.gc_stat = 'GC = {} : GC content = {}'.format(str(self.counter_gc), str(round(stat_gc, 5)))
self.n_stat = 'N = {} '.format(str(self.counter_cn))
self.stats_full = [[i + '\n' for i in n_counts_lst], '\n', self.at_stat, self.gc_stat]
#reset the select seq text box and side bar stats text box
self.side_bar_text_input.setText('')
self.select_seq_text_box.setText('')
stats_lst = [self.side_bar_text_1, self.side_bar_text_2,
self.side_bar_text_3, self.seq_selected, '\n', self.n_counts_header,
self.n_counts_lst_str, '\n', self.at_stat,
'\n', self.gc_stat, '\n\n',
'**Note: N values are dropped in sequence display']
for i in stats_lst:
self.side_bar_text_input.insertPlainText(i)
self.action_message_text_input.setText('Sequence selected.')
except:
try:
if self.data == False:
dlg = load_a_fasta_alert(self)
self.select_seq_text_box.setText('')
else:
dlg = reselect_seq_alert(self)
self.select_seq_text_box.setText('')
except:
pass
def show_seq_data_by_50(self):
'''
Function intended to print the raw string of seq selected to kivy output. It displays
the DNA sequence by 50. An exception is raised if a .fasta file is not
yet loaded or a sequence has not been selected.
[Works as intended]
'''
#at this point self.file_data is the input file as a raw string
try:
self.seq_main_text_input.moveCursor(1, 0)
if (self.seq_selected) and (self.seq_selected in self.seq_keys_nums):
file_data_2 = self.data.split('>')[1:] #converts file_data to list of lists
#REMOVE Ns HERE
table = str.maketrans('', '', 'N')
file_data_2 = [s.translate(table) for s in file_data_2]
seq_selected_idx = int(self.seq_selected)
#format the text...
#file_data_2[seq_selected_idx] is string data of the sequence selected
scoped_data = file_data_2[seq_selected_idx].split('\n')[0:] #creates a list of all data split by \n
self.heading_title = scoped_data[0]
nucleotide_data = ''.join(map(str,scoped_data[1:]))
heading_1_nums = [i for i in range(1,11)]
heading_2_nums = '1234567890'
self.nucleotide_data = '\n'.join(nucleotide_data[i:i + 50] for i in range(0,len(nucleotide_data), 50))
self.nucleotide_data = self.nucleotide_data.split('\n')
idx = 0
line = 1
spaces = ' '
#loop to print each string in the list of strings nucleotides
if self.spacer_tog_checkbox.checkState() == 0:
heading_1_of_5_ns = ' {0[0]} {0[1]} {0[2]} {0[3]} {0[4]}'.format(heading_1_nums)
heading_2_of_5_ns = 'Line {0}{0}{0}{0}{0}'.format(heading_2_nums)
header = ['\n\n' , self.heading_title , ' [Seq Num {}]'.format(self.seq_selected),
'\n', ' ', heading_1_of_5_ns, '\n', heading_2_of_5_ns ]
for i in header:
self.seq_main_text_input.insertPlainText(i)
line_idx = 1
spaces = ' '
for i in self.nucleotide_data:
#self.seq_main_text_input.text += '\n' + spaces + str(line_idx) + ' ' + i
self.seq_main_text_input.insertPlainText('\n')
self.seq_main_text_input.insertPlainText(spaces)
self.seq_main_text_input.insertPlainText(str(line_idx))
self.seq_main_text_input.insertPlainText(' ')
self.seq_main_text_input.insertPlainText(i)
line_idx += 1
if line_idx > 9:
spaces = ' '
elif self.spacer_tog_checkbox.checkState() == 2:
heading_1_of_5_ws = ' {0[0]} {0[1]} {0[2]} {0[3]} {0[4]}'.format(heading_1_nums)
heading_2_of_5_ws = 'Line {0} {0} {0} {0} {0}'.format(heading_2_nums)
header = ['\n\n' , self.heading_title , ' [Seq Num {}]'.format(self.seq_selected),
'\n', ' ', heading_1_of_5_ws, '\n', heading_2_of_5_ws ]
for i in header:
self.seq_main_text_input.insertPlainText(i)
for i in self.nucleotide_data:
inserter_lst = ['\n', spaces, str(line), ' ' , ' '.join(self.nucleotide_data[idx][s:s + 10] for s in range(0, len(self.nucleotide_data[idx]), 10))]
for i in inserter_lst:
self.seq_main_text_input.insertPlainText(i)
idx += 1
line += 1
if line > 9:
spaces = ' '
else:
self.action_message_text_input.setText('Sequence {} loaded.'.format(self.seq_selected))
try:
if (self.seq_selected == False) | (self.seq_selected == ''):
self.action_message_text_input.setText('You need to select a sequence')
except:
self.action_message_text_input.setText('You need to load a fasta file first.')
except:
try:
if self.data:
self.action_message_text_input.setText('You need to select a sequence.')
except:
self.action_message_text_input.setText('You need to load a fasta file first.')
def show_seq_data_by_100(self):
'''
Intended to print the raw string of seq selected to kivy output. It displays
the DNA sequence by 100. An exception is raised if a .fasta file is not
yet loaded or a sequence has not been selected.
[Works as intended]
'''
try:
self.seq_main_text_input.moveCursor(1, 0)
if (self.seq_selected) and (self.seq_selected in self.seq_keys_nums):
file_data_2 = self.data.split('>')[1:] #converts file_data to list of lists
#REMOVE Ns HERE
table = str.maketrans('', '', 'N')
file_data_2 = [s.translate(table) for s in file_data_2]
seq_selected_idx = int(self.seq_selected)
#format the text...
#file_data_2[seq_selected_idx] is string data of the sequence selected
scoped_data = file_data_2[seq_selected_idx].split('\n')[0:] #creates a list of all data split by \n
self.heading_title = scoped_data[0]
nucleotide_data = ''.join(map(str,scoped_data[1:]))
heading_1_nums = [i for i in range(1,11)]
heading_2_nums = '1234567890'
#self.nucleotide_data = '\n 1 '.join(nucleotide_data[i:i + 50] for i in range(0,len(nucleotide_data), 50))
self.nucleotide_data = '\n'.join(nucleotide_data[i:i + 100] for i in range(0,len(nucleotide_data), 100))
self.nucleotide_data = self.nucleotide_data.split('\n')
idx = 0
line = 1
spaces = ' '
#loop to print each string in the list of strings nucleotides
if self.spacer_tog_checkbox.checkState() == 0:
heading_1_of_10_ns = ' {0[0]} {0[1]} {0[2]} {0[3]} {0[4]} {0[5]} {0[6]} {0[7]} {0[8]} {0[9]}'.format(heading_1_nums)
heading_2_of_10_ns = 'Line {0}{0}{0}{0}{0}{0}{0}{0}{0}{0}'.format(heading_2_nums)
header = ['\n\n' , self.heading_title , ' [Seq Num {}]'.format(self.seq_selected),
'\n', ' ', heading_1_of_10_ns, '\n', heading_2_of_10_ns ]
for i in header:
self.seq_main_text_input.insertPlainText(i)
line_idx = 1
spaces = ' '
for i in self.nucleotide_data:
self.seq_main_text_input.insertPlainText('\n')
self.seq_main_text_input.insertPlainText(spaces)
self.seq_main_text_input.insertPlainText(str(line_idx))
self.seq_main_text_input.insertPlainText(' ')
self.seq_main_text_input.insertPlainText(i)
line_idx += 1
if line_idx > 9:
spaces = ' '
elif self.spacer_tog_checkbox.checkState() == 2:
heading_1_of_10_ws = ' {0[0]} {0[1]} {0[2]} {0[3]} {0[4]} {0[5]} {0[6]} {0[7]} {0[8]} {0[9]}'.format(heading_1_nums)
heading_2_of_10_ws = 'Line {0} {0} {0} {0} {0} {0} {0} {0} {0} {0}'.format(heading_2_nums)
inserter_lst = ['\n\n', self.heading_title, ' [Seq Num {}]'.format(self.seq_selected),
'\n', ' ', heading_1_of_10_ws, '\n', heading_2_of_10_ws]
for i in inserter_lst:
self.seq_main_text_input.insertPlainText(i)
insterter_lst_data = ['\n', spaces, str(line), ' ' , ' '.join(self.nucleotide_data[idx][s:s + 10] for s in range(0, len(self.nucleotide_data[idx]), 10))]
for i in self.nucleotide_data:
inserter_lst = ['\n', spaces, str(line), ' ' , ' '.join(self.nucleotide_data[idx][s:s + 10] for s in range(0, len(self.nucleotide_data[idx]), 10))]
for i in inserter_lst:
self.seq_main_text_input.insertPlainText(i)
idx += 1
line += 1
if line > 9:
spaces = ' '
except:
try:
if self.data:
self.action_message_text_input.setText('You need to select a sequence.')
except:
self.action_message_text_input.setText('You need to load a fasta file first.')
def plot_gc(self):
'''
Function that generates a bar plot of all the GC nucleotide data in the
sequence that is selected.
'''
try:
if (self.seq_selected) and (self.seq_selected in self.seq_keys_nums):
#step 1: split each self.original_data_lists into 10 equal segments
org_len_lst = [len(i) for i in self.data_vals]
self.org_len_div_10 = [int(i/10) for i in org_len_lst]
#access the list within the list of lists self.original_data_lists and apply org_len_lst/10
#plot_lists = [i/10 for i in org_len_lst]
spliter_idx = 0
self.org_data_by_10 = []
for items_in_lst in self.data_vals:
new_list = ','.join(items_in_lst[s:s + self.org_len_div_10[spliter_idx]] for s in range(0,len(items_in_lst), self.org_len_div_10[spliter_idx]))
new_list = new_list.split(',')
self.org_data_by_10.append(new_list)
spliter_idx += 1
#Count GC Content in each segment of target seq
self.count_gc_per_fin = []
temp_idx = 0
for i in self.org_data_by_10[int(self.seq_selected)]:
count_gc_per = i.count('GC')
self.count_gc_per_fin.append(count_gc_per)
temp_idx += 1
self.gen_idx_lst = []
for i in range(len(self.count_gc_per_fin)):
self.gen_idx_lst.append(i)
self.data_to_df = pd.DataFrame({'seq': self.gen_idx_lst,
'gc_count': self.count_gc_per_fin})
self.data_to_df['proportion'] = self.data_to_df['gc_count'] / self.data_to_df['gc_count'].sum()
#Bin each self.original_data_lists into 3 or 5
#xaxis = gc content ; yaxis = count of segments per bin
#Plot the data:
fig, ax = plt.subplots(1,1, figsize=(6,6), dpi=100)
ax.hist(x=self.data_to_df['proportion'],
bins=5,
edgecolor="r",
alpha=1)
plt.xticks(rotation = 75)
x_label = 'GC Content For Sequence {0} : \n {1}'.format(self.seq_selected , self.data_keys[int(self.seq_selected)])
ax.set_xlabel('Value of GC content')
ax.set_ylabel('Counts of Segments in Each Bin')
ax.set_title(x_label)
plt.show()
else:
try:
if self.data:
self.action_message_text_input.setText('You need to select a sequence.')
except:
self.action_message_text_input.setText('You need to load a fasta file first.')
except:
try:
if self.data:
self.action_message_text_input.setText('You need to select a sequence.')
except:
self.action_message_text_input.setText('You need to load a fasta file first.')
#ASSIGN THE BUTTUN THE FUNCTION EVENT #STEP 10
#note; I'm not sure yet why this works--but it does. It fixes the bool error commented below
#I think it may have something to do with how the layouts or widgets access each other in the Qt module
#AttributeError: 'bool' object has no attribute 'left_layout_textedit'
self.load_fasta_button.clicked.connect(lambda checked: load_file(self))
self.select_seq_button.clicked.connect(lambda checked: select_seq(self))
self.select_seq_button.clicked.connect(lambda checked: select_seq_ana(self))
self.show_seq_data_by_50.clicked.connect(lambda checked: show_seq_data_by_50(self))
self.show_seq_data_by_100.clicked.connect(lambda checked: show_seq_data_by_100(self))
self.plot_gc_button.clicked.connect(lambda checked: plot_gc(self))
#ADD THE MAIN WINDOWS OUTER LAYOUT (MASTER LAYOUT) #STEP 2
self.setLayout(self.outerLayout)
#SET WINDOW TITLE
self.setWindowTitle('ANG DNA Parser')
#SET DEFUALT MINIMUM SIZE OF WINDOW
self.setMinimumSize(1600,900)
if __name__ == "__main__":
app = QApplication(sys.argv)
app.setStyle('Fusion')
window = WindowClassOne()
window.show()
sys.exit(app.exec_())